The core algorithm of Lumi is a variance-stabilizing transformation. The major idea is that measurements at lower intensities are unreliable, and thus their fold changes (for example, 50/25= 2 fold, where the noises are around 20) should be discounted comparing to measurements at high intensity (say, 40,000 / 20,000 = 2 fold, where the noises are around 1,000).
This is very much intuitive. In a sense, it is similar to the previous approach of VSN (variance-stabilizing normalization) by Dr. Huber at the EBI. We will discuss their differences in the following section.
Does the lumi transformation work?
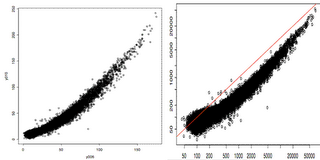
In the figure above, we plotted two technical replicates after lumi transformation (left) and log2 transformation (right). As we expected, the log2 transformation over-inflated the noise at the lower end. Note that the normalization step will take care of the curvature.
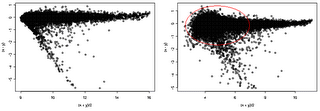
Here we provide additional assessment of the lumi transformation for a pair of treated vs. control arrays. One the left side, we have MvA (difference vs. average) plot of lumi transformed and normalized data. The VSN algorithm (right side) did not adequately stabilize the variance, as evidenced by the larger variance at the low intensity end.
Here is a summary of comparing different data transformation algorithms.